| Twister-sister | |
|---|---|
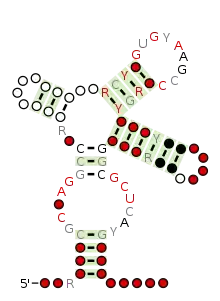 Consensus secondary structure and sequence conservation of Twister_sister_ribozyme | |
| Identifiers | |
| Symbol | Twister-sister |
| Rfam | RF02681 |
| Other data | |
| RNA type | Gene; Ribozyme |
| GO | GO:0003824 |
| SO | SO:0000374 |
| PDB structures | PDBe |
The twister sister ribozyme (TS) is an RNA structure that catalyzes its own cleavage at a specific site. In other words, it is a self-cleaving ribozyme. The twister sister ribozyme was discovered by a bioinformatics strategy [1] as an RNA Associated with Genes Associated with Twister and Hammerhead ribozymes, or RAGATH.
The twister sister ribozyme has a possible structural similarity to twister ribozymes.[1] Some striking similarities were noted, but also surprising differences, such as the absence of the two pseudoknot interactions in the twister ribozyme.[1] The exact nature of the structural relationship between twister and twister sister ribozymes, if any, has not be determined.
Discovery
The twister sister ribozyme was discovered through a bioinformatic search.[1] This study conducted a search for conserved RNA structures near known twister and hammerhead ribozymes as well as certain protein-coding genes based on the fact that many ribozymes are located near to each other and near those genetic fragments. Later they tested the self-cleaving activity of 15 conserved RNA motifs that were found in these regions. 3 out of the 15 RNA motifs showed self-cleaving activity, which were the twister sister ribozyme, the pistol ribozyme and the hatchet ribozyme.[1]
Structure
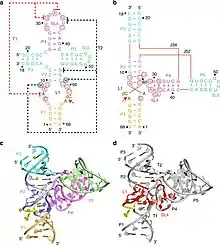
The crystal structures of the pre-catalytic state of the twister sister ribozymes were solved by two research groups independently.
The structure of a three-way junctional twister sister ribozyme is composed of two co-axial stacked helical sections connected with a three-way junction and two tertiary contacts.[2] The active site, a scissile phosphate, is located in a loop with quasihelical character in one coaxial base-stacked helix. Five divalent metal ions are coordinate to RNA ligands, one of which is directly bound to C54 O2’ near the scissile phosphate and exchange inner sphere water molecules with the RNA ligands.[2]
The crystal structure of a four-way junctional twister sister ribozyme is different from the three-way junctional one in terms of long-range interaction and active site structure.[3] The active site of a four-way junctional twister sister is splayed-apart with an interaction between guanine and scissile phosphate. Besides, there are seven divalent metal ions in this ribozyme.[3]
So far, we only know the pre-catalytic conformation of twister sister ribozymes. Understanding the transition state is needed to explain the relationship between twister ribozyme and twister sister ribozyme as well as the structure differences of the active site between the three-way and four-way junctional twister sister ribozymes.
Catalytic mechanism
Generally, nucleolytic ribozymes cleave a specific phosphodiester linkage by SN2 mechanism. The O2' acts as a nucleophile to attack the adjacent P, with O5’ as a leaving group. The catalytic products are a cyclic 2’,3’ phosphate and a 5’-hydroxyl.[4]
The catalytic activity of twister sister increases with pH and depends on divalent metal ion. The cleavage speed increases 10 fold with each increase in pH unit and reach a plateau near pH 7,[2] which indicates that the 2-hydroxyl group of cytidine near the active site is fully deprotonated at pH 7 in the ribozyme. However, the structural basis for the catalytic activity is still under investigation.
The three-way junctional twister sister is a metalloenzyme. The inner sphere water of a divalent metal ion bound to C54 O2’ acts as a general base to deprotonate the 2-hydroxyl group, making it a stronger nucleophile, but the general acid which can stabilize the oxyanion leaving group remains unknown.[2] This mechanism is supported by the exponential correlation between catalytic activity and the pKa of hydrated metal ion.
For the four-way junctional twister sister, Ren and coworkers find that guanine with an amino group is likely to play a role in the catalysis because G5 mutations result in very low catalytic activity. However, it remains unclear whether guanine directly participates in the catalysis as it is not absolutely conserved. The formation of a pseudoknot for four-way junctional TS was found to be highly Mg2+ dependent by conducting SHAPE (Selective-2′ -Hydroxyl Acylation analyzed by Primer Extension) experiments.[5]
References
- 1 2 3 4 5 Weinberg Z, Kim PB, Chen TH, Li S, Harris KA, Lünse CE, Breaker RR (August 2015). "New classes of self-cleaving ribozymes revealed by comparative genomics analysis". Nature Chemical Biology. 11 (8): 606–10. doi:10.1038/nchembio.1846. PMC 4509812. PMID 26167874.
- 1 2 3 4 Liu Y, Wilson TJ, Lilley DM (May 2017). "The structure of a nucleolytic ribozyme that employs a catalytic metal ion". Nature Chemical Biology. 13 (5): 508–513. doi:10.1038/nchembio.2333. PMC 5392355. PMID 28263963.
- 1 2 Zheng L, Mairhofer E, Teplova M, Zhang Y, Ma J, Patel DJ, Micura R, Ren A (October 2017). "Structure-based insights into self-cleavage by a four-way junctional twister-sister ribozyme". Nature Communications. 8 (1): 1180. doi:10.1038/s41467-017-01276-y. PMC 5660989. PMID 29081514.
- ↑ Ren A, Micura R, Patel DJ (December 2017). "Structure-based mechanistic insights into catalysis by small self-cleaving ribozymes". Current Opinion in Chemical Biology. 41: 71–83. doi:10.1016/j.cbpa.2017.09.017. PMC 7955703. PMID 29107885.
- ↑ Gasser C, Gebetsberger J, Gebetsberger M, Micura R (August 2018). "SHAPE probing pictures Mg2+-dependent folding of small self-cleaving ribozymes". Nucleic Acids Research. 46 (14): 6983–6995. doi:10.1093/nar/gky555. PMC 6101554. PMID 29924364.