| Parhyale hawaiensis | |
|---|---|
 | |
| Adult male | |
 | |
| Feeding on a slice of carrot | |
| Scientific classification | |
| Kingdom: | |
| Phylum: | |
| Subphylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | |
| Species: | P. hawaiensis |
| Binomial name | |
| Parhyale hawaiensis Dana, 1853 | |
Parhyale hawaiensis is an amphipod crustacean species that is used in developmental and genetic analyses. It is categorized as an emerging model organism as the main biological techniques necessary for the study of an organism have been established.
Habitat
P. hawaiensis is a detritovore that has a circumtropical, worldwide, intertidal, and shallow-water marine distribution,[1][2] and it may occur as a species complex.[3] It has been reported to occur in large populations (more than 3,000 per square metre) on decaying mangrove leaf material in environments subjected to rapid changes in salinity.[4] The ability to tolerate rapid temperature and osmotic changes allows this species to thrive under typical laboratory conditions. It is a robust species and is being considered for aquaculture.[5]
Life cycle
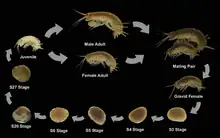
Females produce embryos every 2 weeks once they reach sexual maturity. Embryogenesis is relatively short, lasting about 10 days at 26 °C (79 °F). Females normally brood the embryos in a ventral brood pouch. Close examination of the embryonic development of P. hawaiensis has produced the most detailed staging system for any crustacean.[7] Complete embryogenesis has been divided into 30 discrete stages, which are readily identifiable in living animals or by means of common molecular markers in fixed specimens. Hatchlings possess a complete complement of segments and appendages which are morphologically similar to those of adult animals.
Genetic research
P. hawaiensis is used in genetic research because eggs and embryos are easily manipulated. Embryos can be rapidly and easily removed from the brood pouch and maintained in seawater. Eggs can be collected and hatched individually, and the mature animals can subsequently be used in pairwise sister-brother or mother-son matings to generate inbred lines.
Fertilized eggs are sufficiently large to perform microinjections[8] and blastomere isolations[9] with relative ease. Developing P. hawaiensis embryos are clear, allowing for both detailed microscopic analyses in situ and the use of fluorescently tagged tracer molecules in live embryos. Early cleavage is holoblastic (total), allowing the fates of individual early cells to be explored through experimental manipulation.[7][8][9]
A transcriptome of P. hawaiensis was initially generated using the pyrosequencing technique.[10] This resource is freely available for download. Subsequent, more complete transcriptomes were generated using Illumina sequencing. The transcriptome data can be used for the creation of in situ probes and other experimental tools. The genome is available from NCBI
The genome of P. hawaiensis is available and was sequenced using Illumina short read sequencing.[11] This first version (V3.0) of the genome was recently superseded by a more contiguous version generated by Chicago sequencing through the Dovetail platform. The new version Phaw_5.0 is available on NCBI.
References
- ↑ Clarence Raymond Shoemaker (1956). "Observations on the amphipod genus Parhyale". Proceedings of the United States National Museum. 106 (3372): 345–358. doi:10.5479/si.00963801.106-3372.345.
- ↑ J. L. Barnard (1965). "Marine Amphipoda of atolls in Micronesia". Proceedings of the United States National Museum. 117 (3516): 459–551. doi:10.5479/si.00963801.117-3516.459.
- ↑ Alan A. Myers (1985). "Shallow-water, coral reef and mangrove Amphipoda (Gammaridea) of Fiji" (PDF). Records of the Australian Museum. Suppl 5: 1–143.
- ↑ S. Poovachiranon, K. Boto & N. Duke (1986). "Food preference studies and ingestion rate measurements of the mangrove amphipod Parhyale hawaiensis (Dana)". Journal of Experimental Marine Biology and Ecology. 98 (1–2): 129–140. doi:10.1016/0022-0981(86)90078-X.
- ↑ Vargas-Abúndez, Jorge Arturo; López-Vázquez, Humberto Ivan; Mascaró, Maite; Martínez-Moreno, Gemma Leticia; Simões, Nuno (2021-02-10). "Marine amphipods as a new live prey for ornamental aquaculture: exploring the potential of Parhyale hawaiensis and Elasmopus pectenicrus". PeerJ. 9: e10840. doi:10.7717/peerj.10840. ISSN 2167-8359. PMC 7881717. PMID 33614288.
- ↑ Kao D, Lai AG, Stamataki E, Rosic S, Konstantinides N, Jarvis E, et al. (2016). "The genome of the crustacean Parhyale hawaiensis, a model for animal development, regeneration, immunity and lignocellulose digestion". eLife. 5. doi:10.7554/eLife.20062. PMC 5111886. PMID 27849518.
- 1 2 William E. Browne; Alivia L. Price; Matthias Gerberding; Nipam H. Patel (2005). "Stages of embryonic development in the amphipod crustacean, Parhyale hawaiensis" (PDF). Genesis. 42 (3): 124–149. doi:10.1002/gene.20145. PMID 15986449. S2CID 18213916.
- 1 2 Matthias Gerberding, William E. Browne & Nipam H. Patel (2002). "Cell lineage analysis of the amphipod crustacean Parhyale hawaiensis". Development. 129 (24): 5789–5901. doi:10.1242/dev.00155. PMID 12421717.
- 1 2 Cassandra G. Extavour (2005). "The fate of isolated blastomeres with respect to germ cell formation in the amphipod crustacean Parhyale hawaiensis". Developmental Biology. 277 (2): 387–402. doi:10.1016/j.ydbio.2004.09.030. PMID 15617682.
- ↑ Victor Zeng; Karina Villanueva; Ben Ewen-Campen; Frederike Alwes; William Browne; Cassandra Extavour (2011). "De novo assembly and characterization of a maternal and developmental transcriptome for the emerging model crustacean Parhyale hawaiensis" (PDF). BMC Genomics. 12: 581. doi:10.1186/1471-2164-12-581. PMC 3282834. PMID 22118449.
- ↑ Kao D, Lai AG, Stamataki E, Rosic S, Konstantinides N, Jarvis E; et al. (2016). "The genome of the crustacean Parhyale hawaiensis, a model for animal development, regeneration, immunity and lignocellulose digestion". eLife. 5. doi:10.7554/eLife.20062. PMC 5111886. PMID 27849518.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
Further reading
- E. Jay Rehm; Roberta L. Hannibal; R. Crystal Chaw; Mario A. Vargas-Vila; Nipam H. Patel (2009). "The crustacean Parhyale hawaiensis: a new model for arthropod development". Cold Spring Harbor Protocols. 2009 (1): pdb.emo114. doi:10.1101/pdb.emo114. PMID 20147009.