| "Candidatus Scalindua" | |
|---|---|
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Planctomycetota |
| Class: | Planctomycetia |
| Order: | Planctomycetales |
| Family: | Brocadiaceae |
| Genus: | "Ca. Scalindua" Schmid et al. 2003 |
| Species | |
|
See text | |
"Candidatus Scalindua" is a bacterial genus, and a proposed member of the order Planctomycetales.[1] These bacteria lack peptidoglycan in their cell wall and have a compartmentalized cytoplasm. They are ammonium oxidizing bacteria found in marine environments.
Introduction
"Candidatus Scalindua" is a bacterial genus, member of the order Planctomycetales. These bacteria lack peptidoglycan in their cell wall and have a compartmentalized cytoplasm.[1] "Candidatus Scalindua" spp. can be further divided into three species: Scalindua brodae, Scalindua wagneri, and Scalindua sorokinii. They are ammonium oxidising bacteria found in marine environments. The genus "Ca. Scalindua" are the most abundant anammox bacteria in marine environments, so they are vital to the Earth's nitrogen cycle.[1]
Metabolism
Members of the proposed genus Scalindua are anaerobic anammox (ammonium oxidizing) bacteria.[2] The ammonium-oxidizing reaction composes a significant part of the global nitrogen cycle; by some estimates it is the cause of up to 50% of total nitrogen turnover in marine environments.[3] It consists of the oxidization of ammonium using nitrite as an electron acceptor (both are fixed nitrogen) and subsequent generation of nitrogen gas:
“NH4+ + NO2− = N2 + 2H2O (ΔG° = -357 kj mol-1)”[4]
This reaction uses nitrite (NO2−) as a terminal electron acceptor to produce nitric oxide (NO), which is then combined with ammonium (NH4+) to produce the intermediate hydrazine (N2H4) and water (H2O). Hydrazine, a very reactive molecule also used for rocket fuel, is then oxidized into nitrogen gas (N2).[5] The half reactions may be represented as:
“NO2− + 2H+ + e− = NO + H2O (E° = +0.38V)
NO + NH4+ + 2H+ + 3e− = N2H4 + H2O (E° = +0.06V)
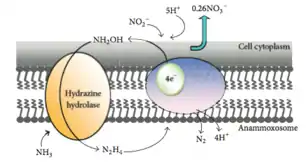
N2H4 = N2 + 4H+ + 4e− (E° = -0.75V)”[4]
This metabolic pathway occurs anaerobically, something that was once considered impossible as ammonium was thought to be inert in the absence of oxygen.[6] In fact, the presence of oxygen over 2 μM inhibits the anammox pathway, which is why members of the proposed genus Scalindua respire anaerobically.[4]
These reactions occur in a large membrane-bound cellular organelle called the anammoxosome, which contains an electron transport chain and an ATPase that pumps protons back into the cytoplasm from the anammoxosome lumen. It functions much like a mitochondrion in eukaryotic cells. The anammoxosome membrane is invaginated (folded in upon itself) to increase its surface area.[7] The existence of membrane-bound cellular organelles is very unusual in prokaryotes, and appears to be limited to the members of the phylum Planctomycetota.[3]
Anammox bacteria, including those belonging to Ca. Scalindua, fix carbon using carbon dioxide as a carbon source. Metagenomic analysis has revealed the presence of genes responsible for the “reductive acetyl-CoA pathway (also known as the Wood-Ljungdahl pathway) which allows for the creation of the precursor molecule acetyl CoA from carbon dioxide.[8][9]
Discovery and distribution
Ammonium and methane are known to be relatively difficult to activate with reactions catalyzed by enzymes that make use of high-potential oxygen radicals, which are unavailable to anaerobic life, leading to the assumption that both compounds were effectively inactive in low oxygen environments.[6] Throughout the 1970s and 80s, results from several independent studies exploring relationships between methane and sulfate concentrations in marine sediments found indications that anaerobic methane oxidation was in fact a widespread occurrence.[6] It was not until 1999 that the existence of anaerobic ammonium oxidation was first discovered in a wastewater treatment plant in The Netherlands and given the name “anammox,” which would later prove to be a key player as part of the marine nitrogen cycle.[6][10][11] Some known anammox bacteria include Candidatus Scalindua, Kuenenia, Brocadia, Jettenia and Anammoxoglobus.[12] Of these bacteria, only Candidatus Scalindua spp. can be found in marine ecosystems.[12]
During the past, many microorganisms such as anammox bacteria may have escaped discovery due to their relatively low growth rates requiring very efficient biomass retention absent from classical methods of cultivation.[13] With the use of biofilms to improve the culturability of organisms that naturally occur in biofilms, combined with the use of biomass retention to study slowly growing microorganisms under substrate limitation, a technique using sequencing batch reactors (SBR) was developed for the long-term enrichment, cultivation, and quantitative analysis of a very slowly growing microbial community.[13] Phylogenetic analysis of the first anammox bacteria discovered concluded that the organisms branched deeply in the phylum, Planctomycetota, which was previously considered to be of limited environmental importance.[10] Nitrogen loses that could only be explained by the process of anammox continued to be discovered in freshwater waste-treatment facilities around the world including North America, Asia, and multiple regions throughout Europe.[14] The role of bacteria belonging to Ca. Scalindua in the marine nitrogen cycle has been found to be of important in the reduction of nitrate to atmospheric nitrogen in anoxic regions of the ocean.[11] Since primary productivity in the ocean is often limited by nitrogen availability, the removal of usable nitrogen in sediments through anammox by Ca. Scalindua may significantly affect biogeochemical cycles in anoxic waters.[11] In certain regions, such as the Golfo Dulce in Costa Rica, up to %35 of atmospheric nitrogen production in the water column can be attributed to Ca. Scalindua spp.[11] In other regions such as the Black Sea, the world's largest anoxic basin, characterized by a large gradient in ammonium concentrations (high levels in deep water tapering off to only trace amounts in the suboxic zone), the apparent ammonium sink in the suboxic zone was identified to be the result of anaerobic oxidation by bacteria belonging to Ca. Scalindua spp.[15]
Morphology
Organisms within the genus “Candidatus Scalindua” are classified as gram-negative chemolithoautotrophic bacteria.[16] This means that their carbon and energy largely come from inorganic sources. Furthermore, bacteria in the genus Ca. Scalindua are obligate anaerobes, so they are unable live in oxygen-rich environments.[16][1]
As with all other organisms within the order Planctomycetota, the cell wall does not contain peptidoglycan.[1][17] The cells are spherical in shape, with a diameter of roughly one micrometer, and contain compartmentalized cytoplasms.[1] Furthermore, organisms within Ca. Scalindua have two inner membranes instead of one inner and one outer membrane surrounding the cell wall.[18] Cells within Ca. Scalindua wagenri are oriented into compact clusters, whereas Ca. Scalindua brodae's cells are more loosely packed.[1] All cells within Ca. Scalindua spp. contain unique organelles called anammoxosomes, which are membrane bound within the cytoplasm.[1][19] Anammoxosomes are where anaerobic ammonium oxidation process occurs. The membrane that surrounds anammoxosomes in anammox bacteria contains unique lipids called “ladderane” lipids, which contain a series of cyclobutane ring structures.[19] However, all other membranes within anammox bacteria are similar to organisms within the order Planctomycetales.
Evolutionary history
According to Strous et al., anammox-capability is the result of a singular evolutionary event. All anammox bacteria are descendants of the same ancient Planctomycetota species that first evolved the anammox reaction.[6] Members of the proposed genus Ca. Scalindua are the most widespread of all the genera of anammox bacteria described so far.[1]
Currently, all anammox bacteria are thought to be members of the order Brocadiales.[20]
Taxonomy
Members belonging to Candidatus Scalindua are close genetic relatives to other anammox bacteria within the order Planctomycetales, such as "Candidatus Brocadia" and "Candidatus Kuenenia".[1] Yet, members of Ca. Scalindua are quite different from other proposed genera of anammox bacteria in terms of their 16S ribosomal RNA sequences.[1] For example, Candidatus Scalindua and Candidatus Brocadia only share 85% similarity in their 16S rRNA sequences.[1] "Candidatus Scalindua" can be further divided into the following three species: "Ca. Scalindua brodae", "Ca. Scalindua wagneri", and "Ca. Scalindua sorokinii".[1][21] Cells belonging to Ca. Scalindua spp. are the most abundant members of Anammox bacteria known to date, making it very important in the world's aquatic environments.
Phylogeny
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN)[22] and National Center for Biotechnology Information (NCBI)[23]
| 120 marker proteins based GTDB 08-RS214[24][25][26] | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Species incertae sedis:
- "Ca. S. erythraensis" Ali et al. 2020
- "Ca. S. flavimaris" corrig. Ahmed et al. 2017
- "Ca. S. marina" van de Vossenberg et al. 2007
- "Ca. S. pacifica" Dang et al. 2013
- "Ca. S. profunda" Van De Vossenberg et al. 2008
- "Ca. S. richardsii" Fuchsman et al. 2012
- "Ca. S. wagneri" Schmid et al. 2003
- "Ca. S. zhenghei" Hong et al. 2011
Ecological role
Candidatus Scalindua sp. is the only taxonomic group of ammonium-oxidizing bacteria found in the Black Sea, the Benguela Oxygen minimum zone off the coast of Namibia, and the estuary of the Randers Fjord, Denmark.[27] Globally, members of Candidatus Scalindua spp. have been discovered in all marine environments that have been studied; most other marine bacteria are not this wide spread.[27][12]
The ideal environmental conditions, with regards to temperature, pH and salinity for “Candidatus Scalindua sp.” are as follows: 10 to 30 °C, 6.0 to 8.5 pH and 0.8% to 4.0% salinity. No ammonium oxidizing activity was observed when salinity was 0%.[28]
Marine sediments located in deep-sea methane seeps contain anammox bacteria associated with Candidatus Scalindua spp.; these bacteria likely have a substantial role in the nitrogen cycle in the sediments.[29]
Two types of anammox bacteria belonging to Ca. Scalindua (59% abundance) and Ca. Kuenenia (41% abundance), have been found in the non-rhizosphere area of the saltmarsh grass Spartina alterniflora while only Ca. Scalindua was present within the rhizosphere. Moreover, it was in 1.5 times greater abundance than for other anammox bacterial in the non-rhizosphere sediments.[30] Changing seasons do not affect the make-up of anammox-capable bacterial communities within the sediments in and around the rhizosphere; however, there was always a greater abundance of anammox bacteria within the rhizosphere that peaked in abundance during July and October when temperatures are warmest.[30] During the warmer parts of the year both communities of anammox bacteria within and outside of the rhizosphere are more active, and produce more N2 with the bacteria in the rhizosphere producing almost twice as much N2.[30]
Applications
Bacteria belonging to "Ca. Scalindua wagneri" are often used in wastewater treatment plants to reduce the adverse effects of nitrification and denitrification on the local environment.[31] The use of anammox bacteria in wastewater treatment plants has a drastically reduced cost compared to previous denitrification methods. Furthermore, it is a much more environmentally friendly method.[8]
Species
References
- 1 2 3 4 5 6 7 8 9 10 11 12 13 Schmid, Markus (2003). "Candidatus "Scalindua brodae", sp. nov., Candidatus "Scalindua wagneri", sp. nov., Two New Species of Anaerobic Ammonium Oxidizing Bacteria". Systematic and Applied Microbiology. 26 (4): 529–538. doi:10.1078/072320203770865837. ISSN 0723-2020. PMID 14666981.
- ↑ Awata, Takanori; Oshiki, Mamoru; Kindaichi, Tomonori; Ozaki, Noriatsu; Ohashi, Akiyoshi; Okabe, Satoshi (2017-03-08). "Physiological Characterization of an Anaerobic Ammonium-Oxidizing Bacterium Belonging to the "Candidatus Scalindua" Group". Applied and Environmental Microbiology. 79 (13): 4145–4148. Bibcode:2013ApEnM..79.4145A. doi:10.1128/AEM.00056-13. ISSN 0099-2240. PMC 3697556. PMID 23584767.
- 1 2 Kuenen, J. Gijs (2008-04-01). "Anammox bacteria: from discovery to application". Nature Reviews Microbiology. 6 (4): 320–326. doi:10.1038/nrmicro1857. ISSN 1740-1526. PMID 18340342. S2CID 6378856.
- 1 2 3 "Anammox Bacteria". Microbewiki.
- ↑ K., Poole, Robert (2012-01-01). Advances in microbial physiology Volume 60. Elsevier/Academic Press. ISBN 9780123982643. OCLC 797831085.
{{cite book}}: CS1 maint: multiple names: authors list (link) - 1 2 3 4 5 Strous, Marc; Jetten, Mike S. M. (2004-09-13). "Anaerobic Oxidation of Methane and Ammonium". Annual Review of Microbiology. 58: 99–117. doi:10.1146/annurev.micro.58.030603.123605. PMID 15487931.
- ↑ Niftrik, Laura van; Jetten, Mike S. M. (2012-09-01). "Anaerobic Ammonium-Oxidizing Bacteria: Unique Microorganisms with Exceptional Properties". Microbiology and Molecular Biology Reviews. 76 (3): 585–596. doi:10.1128/MMBR.05025-11. ISSN 1092-2172. PMC 3429623. PMID 22933561.
- 1 2 van de Vossenberg, Jack; Woebken, Dagmar; Maalcke, Wouter J; Wessels, Hans J C T; Dutilh, Bas E; Kartal, Boran; Janssen-Megens, Eva M; Roeselers, Guus; Yan, Jia (2017-03-08). "The metagenome of the marine anammox bacterium 'Candidatus Scalindua profunda' illustrates the versatility of this globally important nitrogen cycle bacterium". Environmental Microbiology. 15 (5): 1275–1289. doi:10.1111/j.1462-2920.2012.02774.x. ISSN 1462-2912. PMC 3655542. PMID 22568606.
- ↑ Kartal, Boran; Almeida, De; M, Naomi; Maalcke, Wouter J.; Camp, Op den; J.m, Huub; Jetten, Mike S. M.; Keltjens, Jan T. (2013-05-01). "How to make a living from anaerobic ammonium oxidation". FEMS Microbiology Reviews. 37 (3): 428–461. doi:10.1111/1574-6976.12014. ISSN 0168-6445. PMID 23210799.
- 1 2 Jetten, Mike S. M.; Strous, Marc; Fuerst, John A.; Kramer, Evelien H. M.; Logemann, Susanne; Muyzer, Gerard; Pas-Schoonen, Katinka T. van de; Webb, Richard; Kuenen, J. Gijs (1999). "Missing lithotroph identified as new planctomycete" (PDF). Nature. 400 (6743): 446–449. Bibcode:1999Natur.400..446S. doi:10.1038/22749. PMID 10440372. S2CID 2222680.
- 1 2 3 4 Dalsgaard, Tage; Canfield, Donald E.; Petersen, Jan; Thamdrup, Bo; Acuña-González, Jenaro (2003). "N2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica". Nature. 422 (6932): 606–608. Bibcode:2003Natur.422..606D. doi:10.1038/nature01526. PMID 12686998. S2CID 4318646.
- 1 2 3 Woebken, Dagmar; Lam, Phyllis; Kuypers, Marcel M. M.; Naqvi, S. Wajih A.; Kartal, Boran; Strous, Marc; Jetten, Mike S. M.; Fuchs, Bernhard M.; Amann, Rudolf (2008-11-01). "A microdiversity study of anammox bacteria reveals a novel Candidatus Scalindua phylotype in marine oxygen minimum zones". Environmental Microbiology. 10 (11): 3106–3119. doi:10.1111/j.1462-2920.2008.01640.x. ISSN 1462-2920. PMID 18510553.
- 1 2 Strous, M.; Heijnen, J. J.; Kuenen, J. G.; Jetten, M. S. M. (1998-11-01). "The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms". Applied Microbiology and Biotechnology. 50 (5): 589–596. doi:10.1007/s002530051340. ISSN 0175-7598. S2CID 33437272.
- ↑ Schmid, Markus C.; Maas, Bart; Dapena, Ana; Pas-Schoonen, Katinka van de; Vossenberg, Jack van de; Kartal, Boran; Niftrik, Laura van; Schmidt, Ingo; Cirpus, Irina (2005-04-01). "Biomarkers for In Situ Detection of Anaerobic Ammonium-Oxidizing (Anammox) Bacteria". Applied and Environmental Microbiology. 71 (4): 1677–1684. Bibcode:2005ApEnM..71.1677S. doi:10.1128/AEM.71.4.1677-1684.2005. ISSN 0099-2240. PMC 1082507. PMID 15811989.
- ↑ Kuypers, Marcel M. M.; Sliekers, A. Olav; Lavik, Gaute; Schmid, Markus; Jørgensen, Bo Barker; Kuenen, J. Gijs; Damsté, Jaap S. Sinninghe; Strous, Marc; Jetten, Mike S. M. (2003). "Anaerobic ammonium oxidation by anammox bacteria in the Black Sea". Nature. 422 (6932): 608–611. Bibcode:2003Natur.422..608K. doi:10.1038/nature01472. PMID 12686999. S2CID 4318175.
- 1 2 Strous, M. (1999). "Missing lithotroph identified as new planctomycete" (PDF). Nature. 400 [6743] (6743): 446–449. Bibcode:1999Natur.400..446S. doi:10.1038/22749. PMID 10440372. S2CID 2222680.
- ↑ Lindsay, M.R (2001). "Cell compartmentalisation in planctomycetes: Bovel types of structural organisation for the bacterial cell". Archives of Microbiology. 175 [6] (6): 413–429. doi:10.1007/s002030100280. PMID 11491082. S2CID 21970703.
- ↑ van Niftrick, Laura A. (2004). "The anammoxosome: an intracytoplasmic compartment in anammox bacteria". FEMS Microbiology Letters. 233 (1): 7–13. doi:10.1016/j.femsle.2004.01.044. PMID 15098544.
- 1 2 Sinninghe, Damste (2002). "Linearly concatenated cyclobutane lipids form a dense bacterial membrane". Nature. 419 (6908): 708–712. Bibcode:2002Natur.419..708S. doi:10.1038/nature01128. PMID 12384695. S2CID 4373854.
- ↑ Speth, Daan R.; Russ, Lina; Kartal, Boran; op den Camp, Huub J. M.; Dutilh, Bas E.; Jetten, Mike S. M. (2015-01-08). "Draft Genome Sequence of Anammox Bacterium "Candidatus Scalindua brodae," Obtained Using Differential Coverage Binning of Sequencing Data from Two Reactor Enrichments". Genome Announcements. 3 (1): e01415–14. doi:10.1128/genomeA.01415-14. ISSN 2169-8287. PMC 4290996. PMID 25573945.
- ↑ Kuypers, M. (2003). "Anaerobic ammonium oxidation by Anammox bacteria in the Black Sea". Nature. 422 (6932): 608–611. Bibcode:2003Natur.422..608K. doi:10.1038/nature01472. PMID 12686999. S2CID 4318175.
- ↑ A.C. Parte; et al. "Scalindua". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved 2022-09-09.
- ↑ Sayers; et al. "Scalindua". National Center for Biotechnology Information (NCBI) taxonomy database. Retrieved 2022-09-09.
- ↑ "GTDB release 08-RS214". Genome Taxonomy Database. Retrieved 10 May 2023.
- ↑ "bac120_r214.sp_label". Genome Taxonomy Database. Retrieved 10 May 2023.
- ↑ "Taxon History". Genome Taxonomy Database. Retrieved 10 May 2023.
- 1 2 Schmid, Markus C.; Risgaard-Petersen, Nils; Van De Vossenberg, Jack; Kuypers, Marcel M. M.; Lavik, Gaute; Petersen, Jan; Hulth, Stefan; Thamdrup, Bo; Canfield, Don (2007-06-01). "Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity". Environmental Microbiology. 9 (6): 1476–1484. doi:10.1111/j.1462-2920.2007.01266.x. ISSN 1462-2920. PMID 17504485.
- ↑ Awata, Takanori; Oshiki, Mamoru; Kindaichi, Tomonori; Ozaki, Noriatsu; Ohashi, Akiyoshi; Okabe, Satoshi (2013-07-01). "Physiological Characterization of an Anaerobic Ammonium-Oxidizing Bacterium Belonging to the "Candidatus Scalindua" Group". Applied and Environmental Microbiology. 79 (13): 4145–4148. Bibcode:2013ApEnM..79.4145A. doi:10.1128/AEM.00056-13. ISSN 0099-2240. PMC 3697556. PMID 23584767.
- ↑ Shao, Sudong; Luan, Xiwu; Dang, Hongyue; Zhou, Haixia; Zhao, Yakun; Liu, Haitao; Zhang, Yunbo; Dai, Lingqing; Ye, Ying (2014-02-01). "Deep-sea methane seep sediments in the Okhotsk Sea sustain diverse and abundant anammox bacteria". FEMS Microbiology Ecology. 87 (2): 503–516. doi:10.1111/1574-6941.12241. ISSN 0168-6496. PMID 24164560.
- 1 2 3 Zheng, Yanling; Hou, Lijun; Liu, Min; Yin, Guoyu; Gao, Juan; Jiang, Xiaofen; Lin, Xianbiao; Li, Xiaofei; Yu, Chendi (2016-09-01). "Community composition and activity of anaerobic ammonium oxidation bacteria in the rhizosphere of salt-marsh grass Spartina alterniflora". Applied Microbiology and Biotechnology. 100 (18): 8203–8212. doi:10.1007/s00253-016-7625-2. ISSN 0175-7598. PMID 27225476. S2CID 17385541.
- ↑ Jetten, Mike (2004). "Biodiversity and application of anaerobic ammonium-oxidizing bacteria". European Symposium on Environmental Biotechnology: 21–26.