| Phosphoribosylamine—glycine ligase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 phosphoribosylamine-glycine ligase monomer (fragment), Human | |||||||||
| Identifiers | |||||||||
| EC no. | 6.3.4.13 | ||||||||
| CAS no. | 9032-01-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Phosphoribosylglycinamide synthetase, N domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 glycinamide ribonucleotide synthetase (gar-syn) from e. coli. | |||||||||
| Identifiers | |||||||||
| Symbol | GARS_N | ||||||||
| Pfam | PF02844 | ||||||||
| InterPro | IPR020562 | ||||||||
| PROSITE | PDOC00164 | ||||||||
| SCOP2 | 1gso / SCOPe / SUPFAM | ||||||||
| |||||||||
| Phosphoribosylglycinamide synthetase, ATP-grasp (A) domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 glycinamide ribonucleotide synthetase (gar-syn) from e. coli. | |||||||||
| Identifiers | |||||||||
| Symbol | GARS_A | ||||||||
| Pfam | PF01071 | ||||||||
| Pfam clan | CL0179 | ||||||||
| InterPro | IPR020561 | ||||||||
| PROSITE | PDOC00164 | ||||||||
| SCOP2 | 1gso / SCOPe / SUPFAM | ||||||||
| |||||||||
| Phosphoribosylglycinamide synthetase, C domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of phosphoribosylamine--glycine ligase (tm1250) from thermotoga maritima at 2.30 a resolution | |||||||||
| Identifiers | |||||||||
| Symbol | GARS_C | ||||||||
| Pfam | PF02843 | ||||||||
| InterPro | IPR020560 | ||||||||
| PROSITE | PDOC00164 | ||||||||
| SCOP2 | 1gso / SCOPe / SUPFAM | ||||||||
| |||||||||
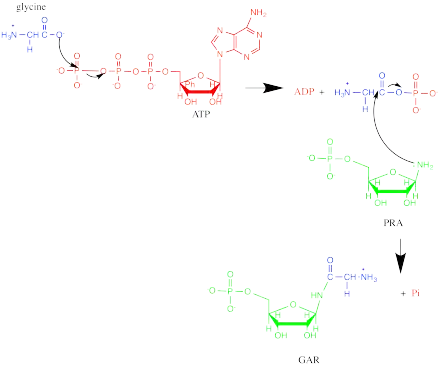
Phosphoribosylamine—glycine ligase, also known as glycinamide ribonucleotide synthetase (GARS), (EC 6.3.4.13) is an enzyme that catalyzes the chemical reaction
- ATP + 5-phospho-D-ribosylamine + glycine ADP + phosphate + N1-(5-phospho-D-ribosyl)glycinamide
which is the second step in purine biosynthesis. The 3 substrates of this enzyme are ATP, 5-phospho-D-ribosylamine, and glycine, whereas its 3 products are ADP, phosphate, and N1-(5-phospho-D-ribosyl)glycinamide.
This enzyme belongs to the family of ligases, specifically those forming generic carbon-nitrogen bonds.
In bacteria, GARS is a monofunctional enzyme (encoded by the purD gene). The purD genes often contain PurD RNA motif in their 5' UTR.[1] In yeast, GARS is part of a bifunctional enzyme (encoded by the ADE5/7 gene) in conjunction with phosphoribosylformylglycinamidine cyclo-ligase (AIRS). In higher eukaryotes, including humans,[2] GARS is part of a trifunctional enzyme in conjunction with AIRS and with phosphoribosylglycinamide formyltransferase (GART),[3] forming GARS-AIRS-GART.
Nomenclature
The systematic name of this enzyme class is 5-phospho-D-ribosylamine:glycine ligase (ADP-forming). Other names in common use include:
- phosphoribosylglycinamide synthetase
- glycinamide ribonucleotide synthetase
- phosphoribosylglycineamide synthetase
- glycineamide ribonucleotide synthetase
- 2-amino-N-ribosylacetamide 5'-phosphate kinosynthase
- 5'-phosphoribosylglycinamide synthetase
- GAR synthetase
Mechanism
GARS operates via an ordered, sequential mechanism. 5-phospho-D-ribosylamine (PRA) binds first, then ATP, and finally glycine. Phosphate is released first, followed by ADP and GAR.[4] The oxygen in the ribose ring of PRA is important in substrate binding, likely due to favorable energetics from hydrogen bonding and the ring conformation it confers.[5] In addition, the phosphate group of GAR has been implicated in GARS substrate recognition. The reaction starts with the oxygen of glycine acting as a nucleophile to attack the γ-phosphorus of ATP. Then, the nitrogen of PRA attacks the carbonyl carbon in the intermediate, and phosphate leaves, forming GAR.[4][6]
Structural studies
As of late 2007, 3 structures have been solved for this class of enzymes, with PDB accession codes 1GSO, 1VKZ, and 2QK4. The overall structure of the enzyme, based on crystallization from E. coli,[7] consists of 16 alpha helices which connect to 20 beta strands by turns and loops. There are four main domains: N, A, B, and C. Each domain has a central beta sheet with an alpha helix on at least one side. The N, A, and C domains are clustered together, while the B domain is slightly separated from the others and connected to them by two hinge regions. The active site is between the NAC group and the B domain. The A and B domains appear to facilitate ATP binding, while the N and C domains confer substrate specificity. The N domain is very similar to that of glycinamide ribonucleotide transformylase. Although the orientation of the B domains varies, the structure of GARS is very similar across organisms.[4] Furthermore, the gene has been sequenced in many organisms, and E. coli shows between 41 and 52% identity with the GARS sequences of B. subtilis, S. cerevisiae, D. melanogaster, and D. pseudobscura.[8] Human GARS-AIRS-GART has been shown to be most similar to that of mice, chimpanzees, and cows.[9] Among the amino acids that are identical in B. subtilis, S. cerevisiae, D. melanogaster, and D. pseudobscura, almost a third are glycine and proline, which suggests that they play an important role in proper folding of the protein.[8] In addition to similar structure across species, GARS as a whole has a very similar structure to D-alanine:D-alanine ligase, biotin carboxylase, and glutathione synthetase. All of these enzymes have an ATP binding domain classified as ATP-grasp domains.[4]
Disease relevance
In humans, the gene that codes for GARS-AIRS-GART is on chromosome 21, and individuals with Down Syndrome have higher purine levels, which has been correlated with mental retardation. Thus, studies have been conducted to investigate its involvement in Down Syndrome. It has been found that GARS is expressed for longer in individuals with Down Syndrome than in unaffected individuals.[10] In unaffected individuals, GARS is highly expressed in the cerebellum before birth but is barely expressed by three weeks after birth. In individuals with Down Syndrome, GARS expression continues until at least seven weeks after birth. This suggests that GARS may be a main contributor to the development of Down Syndrome. However, so far no mutations to GARS have been identified that could change its function and cause Down Syndrome related mental retardation.[11]
References
- ↑ Weinberg Z, Barrick JE, Yao Z, et al. (2007). "Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline". Nucleic Acids Res. 35 (14): 4809–19. doi:10.1093/nar/gkm487. PMC 1950547. PMID 17621584.
- ↑ Daubner, Susan Colette (13 January 1986). "Structural and Mechanistic Studies on the HeLa and Chicken Liver Proteins That Catalyze Glycinamide Ribonucleotide Synthesis and Formylation and Aminoimidazole Ribonucleotide Synthesis". Biochemistry. 25 (10): 2953–2957. doi:10.1021/bi00358a033. PMID 3718932.
- ↑ Daubner, Susan Colette (5 August 1985). "A Multifunctional Protein Possessing Glycinamide Ribonucleotide Synthetase, Glycinamide Ribonucleotide Transformylase, and Aminoimidazole Ribonucleotide Synthetase Activities in de Novo Purine Biosynthesis". Biochemistry. 24 (25): 7059, 7061–7062. doi:10.1021/bi00346a006. PMID 4084560.
- 1 2 3 4 Sampei, Gen-ichi (16 August 2010). "Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria". Journal of Biochemistry. 148 (4): 429–437. doi:10.1093/jb/mvq088. PMID 20716513.
- ↑ Antle, Vincent D. (5 April 1996). "Substrate Specificity of Glycinamide Ribonucleotide Synthetase from Chicken Liver". The Journal of Biological Chemistry. 271 (14): 8194–8195. doi:10.1074/jbc.271.14.8192. PMID 8626510.
- ↑ Kappock, T Joseph (1 October 2000). "Modular evolution of the purine biosynthetic pathway". Current Opinion in Chemical Biology. 4 (5): 567–72. doi:10.1016/S1367-5931(00)00133-2. PMID 11006546.
- ↑ Wang, Weiru (1998). "X-Ray Crystal Structure of Glycinamide Ribonucleotide Synthetase from Escherichia coli". Biochemistry. 37 (45): 15647–15648, 15651–15652. doi:10.1021/bi981405n. PMID 9843369.
- 1 2 Aiba, Atsu (12 April 1989). "Nucleotide Sequence Analysis of Genes purH and purD Involved in the de Novo Purine Biosynthesis of Escherchia coli". The Journal of Biological Chemistry. 264 (35): 21239–46. PMID 2687276. Retrieved 8 March 2015.
- ↑ Banerjee, Disha (20 March 2009). "Phylogenetic Analysis and in Silico Characterization of the GARS-AIRS-GART Gene which Codes for a tri-Functional Enzyme Protein Involved in de Novo Purine Biosynthesis". Molecular Biotechnology. 42 (3): 306–317. doi:10.1007/s12033-009-9160-1. PMID 19301155. S2CID 34759623.
- ↑ Brodsky, Gary (9 August 1997). "The human GARS-AIRS-GART gene encodes two proteins which are differentially expressed during human brain development and temporally overexpressed in cerebellum of individuals with Down syndrome". Human Molecular Genetics. 6 (12): 2043–2044, 2046–2048. doi:10.1093/hmg/6.12.2043. PMID 9328467.
- ↑ Banerjee, Disha (10 November 2011). "No Evidence for Mutations that Deregulate GARS-AIRS-GART Protein Levels in Children with Down Syndrome" (PDF). Indian Journal of Clinical Biochemistry. 27 (1): 46–50. doi:10.1007/s12291-011-0183-6. PMC 3286581. PMID 23277712. Retrieved 8 March 2015.