| Myosin Light-Chain kinase, smooth muscle | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | MYLK | ||||||
| NCBI gene | 4638 | ||||||
| HGNC | 7590 | ||||||
| OMIM | 600922 | ||||||
| RefSeq | NM_053025 | ||||||
| UniProt | Q15746 | ||||||
| Other data | |||||||
| EC number | 2.7.11.18 | ||||||
| Locus | Chr. 3 qcen-q21 | ||||||
| |||||||
| myosin light-chain kinase 2, skeletal muscle | |||||||
|---|---|---|---|---|---|---|---|
 | |||||||
| Identifiers | |||||||
| Symbol | MYLK2 | ||||||
| NCBI gene | 85366 | ||||||
| HGNC | 16243 | ||||||
| OMIM | 606566 | ||||||
| RefSeq | NM_033118 | ||||||
| UniProt | Q9H1R3 | ||||||
| Other data | |||||||
| Locus | Chr. 20 q13.31 | ||||||
| |||||||
| myosin light-chain kinase 3, cardiac | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | MYLK3 | ||||||
| NCBI gene | 91807 | ||||||
| HGNC | 29826 | ||||||
| OMIM | 612147 | ||||||
| RefSeq | NM_182493 | ||||||
| UniProt | Q32MK0 | ||||||
| Other data | |||||||
| Locus | Chr. 16 q11.2 | ||||||
| |||||||
| Human Myosin Light-Chain Kinase | |||||||
|---|---|---|---|---|---|---|---|
 The Crystal Structure of the Human Myosin Light Chain Kinase Loc340156.[2] | |||||||
| Identifiers | |||||||
| Symbol | MYLK4 | ||||||
| NCBI gene | 340156 | ||||||
| HGNC | 27972 | ||||||
| RefSeq | NM_001012418 | ||||||
| UniProt | Q86YV6 | ||||||
| |||||||
Myosin light-chain kinase also known as MYLK or MLCK is a serine/threonine-specific protein kinase that phosphorylates a specific myosin light chain, namely, the regulatory light chain of myosin II.[3]
General structural features
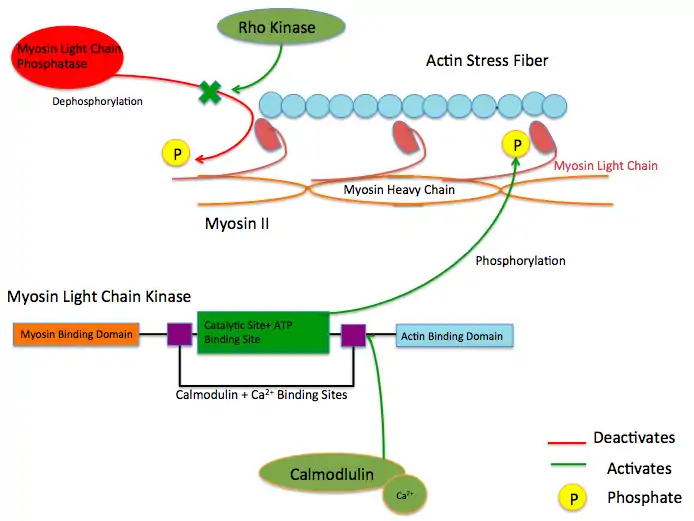
While there are numerous differing domains depending on the cell type, there are several characteristic domains common amongst all MYLK isoforms. MYLK’s contain a catalytic core domain with an ATP binding domain. On either sides of the catalytic core sit calcium ion/calmodulin binding sites. Binding of calcium ion to this domain increases the affinity of MYLK binding to myosin light chain. This myosin binding domain is located at the C-Terminus end of the kinase. On the other side of the kinase at the N-Terminus end, sits the actin-binding domain, which allows MYLK to form interactions with actin filaments, keeping it in place.[4][5]
Isoforms
Four different MYLK isoforms exist:[6]
Function
These enzymes are important in the mechanism of contraction in muscle. Once there is an influx of calcium cations (Ca2+) into the muscle, either from the sarcoplasmic reticulum or from the extracellular space, contraction of smooth muscle fibres may begin. First, the calcium will bind to calmodulin.[7] After the influx of calcium ions and the binding to calmodulin, pp60 SRC (a protein kinase) causes a conformational change in MYLK, activating it and resulting in an increase in phosphorylation of myosin light chain at serine residue 19. The phosphorylation of MLC will enable the myosin crossbridge to bind to the actin filament and allow contraction to begin (through the crossbridge cycle). Since smooth muscle does not contain a troponin complex, as striated muscle does, this mechanism is the main pathway for regulating smooth muscle contraction. Reducing intracellular calcium concentration inactivates MLCK but does not stop smooth muscle contraction since the myosin light chain has been physically modified through phosphorylation(and not via ATPase activity). To stop smooth muscle contraction this change needs to be reversed. Dephosphorylation of the myosin light chain (and subsequent termination of muscle contraction) occurs through activity of a second enzyme known as myosin light-chain phosphatase (MLCP).[8]
Upstream Regulators
Protein kinase C and ROC Kinase are involved in regulating Calcium ion intake; these Calcium ions, in turn stimulate a MYLK, forcing a contraction.[9] Rho kinase also modulates the activity of MYLK by downregulating the activity of MYLK's counterpart protein: Myosin Light Chain Phosphatase (MYLP).[10] In addition to downregulation of MYLK, ROCK indirectly strengthens actin/myosin contraction through inhibiting Cofilin, a protein which depolymerizes actin stress fibers.[11] Similar to ROCK, Protein Kinase C regulates MYLK via the CPI-17 protein, which downregulates MYLP.[12]
Mutations and resulting diseases
Some pulmonary disorders have been found to arise due to an inability of MYLK to function properly in lung cells. Over-activity in MYLK creates an imbalance in mechanical forces between adjacent endothelial and lung tissue cells. An imbalance may result in acute respiratory distress syndrome, in which fluid is able to pass into the alveoli.[13] Within the cells, MYLK provides an inward pulling force, phosphorylating myosin light chain causing a contraction of the myosin/actin stress fiber complex. Conversely, cell-cell adhesion via tight and adherens junctions, along with anchoring to extra cellular matrix (ECM) via integrins and focal adhesion proteins results in an outward pulling force. Myosin light chain pulls the actin stress fiber attached to the cadherin, resisting the force of the adjacent cell's cadherin. However, when the inward pulling force of the actin stress fiber becomes greater than the outward pulling force of the cell adhesion molecules due to an overactive MYLK, tissues can become slightly pulled apart and leaky, leading to passage of fluid into the lungs.[14]
Another source of smooth muscle disorders like ischemia–reperfusion, hypertension, and coronary artery disease arise when mutations to protein kinase C (PKC) result in excessive inhibition of MYLP, which counteracts the activity of MYLK by dephosphorylating myosin light chain. Because myosin light chain has no inherent phosphate cleaving property over active PKC prevents the dephosphorylation of myosin light protein leaving it in the activated conformation, causing an increase in smooth muscle contraction.[12]
See also
References
- ↑ Radu, L.; Assairi, L.; Blouquit, Y.; Durand, D.; Miron, S.; Charbonnier, J.B.; Craescu, C.T. (2011). "RCSB Protein Data Bank - Structure Summary for 3KF9 - Crystal structure of the SdCen/skMLCK complex". Worldwide Protein Data Bank. doi:10.2210/pdb3kf9/pdb.
- ↑ Muniz, J.R.C.; Mahajan, P.; Rellos, P.; Fedorov, O.; Shrestha, B.; Wang, J.; Elkins, J.M.; Daga, N.; Cocking, R.; Chaikuad, A.; Krojer, T.; Ugochukwu, E.; Yue, W.; von Delft, F.; Arrowsmith, C.H.; Edwards, A.M.; Weigelt, J.; Bountra, C.; Gileadi, O.; Knapp, S. (2010). "RCSB Protein Data Bank - Structure Summary for 2X4F - The Crystal Structure of the Human Myosin Light Chain Kinase Loc340156". Worldwide Protein Data Bank. doi:10.2210/pdb2x4f/pdb.
- ↑ Gao Y, Ye LH, Kishi H, Okagaki T, Samizo K, Nakamura A, Kohama K (June 2001). "Myosin light chain kinase as a multifunctional regulatory protein of smooth muscle contraction". IUBMB Life. 51 (6): 337–44. doi:10.1080/152165401753366087. PMID 11758800. S2CID 46180993.
- ↑ Khapchaev AY, Shirinsky VP (December 2016). "Myosin Light Chain Kinase MYLK1: Anatomy, Interactions, Functions, and Regulation". Biochemistry. Biokhimiia. 81 (13): 1676–1697. doi:10.1134/S000629791613006X. PMID 28260490. S2CID 11424747.
- ↑ Stull JT, Lin PJ, Krueger JK, Trewhella J, Zhi G (December 1998). "Myosin Light Chain Kinase: Functional Domains and Structural Motifs". Acta Physiologica. 164 (4): 471–482. doi:10.1111/j.1365-201X.1998.tb10699.x. PMID 9887970.
- ↑ Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (December 2002). "The protein kinase complement of the human genome". Science. 298 (5600): 1912–34. Bibcode:2002Sci...298.1912M. doi:10.1126/science.1075762. PMID 12471243. S2CID 26554314.
- ↑ Robinson A, Colbran R (2013). "Calcium/Calmodulin-Dependent Protein Kinases". In Lennarz W, Lane D (eds.). Encyclopedia of Biological Chemistry (2nd ed.). Elsevier inc. pp. 304–309. ISBN 978-0-12-378631-9.
- ↑ Feher J (2017). "Smooth Muscle". Quantitative Human Physiology (2nd ed.). Elsevier inc. pp. 351–361. ISBN 978-0-12-800883-6.
- ↑ Anjum I (January 2018). "Calcium sensitization mechanisms in detrusor smooth muscles". Journal of Basic and Clinical Physiology and Pharmacology. 29 (3): 227–235. doi:10.1515/jbcpp-2017-0071. PMID 29306925. S2CID 20486807.
- ↑ Amano M, Nakayama M, Kaibuchi K (September 2010). "Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell polarity". Cytoskeleton. 67 (9): 545–54. doi:10.1002/cm.20472. PMC 3038199. PMID 20803696.
- ↑ Dudek SM, Garcia JG (October 2001). "Cytoskeletal regulation of pulmonary vascular permeability". Journal of Applied Physiology. 91 (4): 1487–500. doi:10.1152/jappl.2001.91.4.1487. PMID 11568129. S2CID 7042112.
- 1 2 Ringvold HC, Khalil RA (2017). "Protein Kinase C as Regulator of Vascular Smooth Muscle Function and Potential Target in Vascular Disorders". Vascular Pharmacology - Smooth Muscle. Advances in Pharmacology. Vol. 78. pp. 203–301. doi:10.1016/bs.apha.2016.06.002. ISBN 978-0-12-811485-8. PMC 5319769. PMID 28212798.
- ↑ Szilágyi KL, Liu C, Zhang X, Wang T, Fortman JD, Zhang W, Garcia JG (February 2017). "Epigenetic contribution of the myosin light chain kinase gene to the risk for acute respiratory distress syndrome". Translational Research. 180: 12–21. doi:10.1016/j.trsl.2016.07.020. PMC 5253100. PMID 27543902.
- ↑ Cunningham KE, Turner JR (July 2012). "Myosin Light Chain Kinase: Pulling the Strings of Epithelial Tight Junction Function". Annals of the New York Academy of Sciences. 1258 (1): 34–42. Bibcode:2012NYASA1258...34C. doi:10.1111/j.1749-6632.2012.06526.x. PMC 3384706. PMID 22731713.
Further reading
- Clayburgh DR, Rosen S, Witkowski ED, Wang F, Blair S, Dudek S, Garcia JG, Alverdy JC, Turner JR (December 2004). "A differentiation-dependent splice variant of myosin light chain kinase, MLCK1, regulates epithelial tight junction permeability". The Journal of Biological Chemistry. 279 (53): 55506–13. doi:10.1074/jbc.M408822200. PMC 1237105. PMID 15507455.
- Wang F, Graham WV, Wang Y, Witkowski ED, Schwarz BT, Turner JR (February 2005). "Interferon-gamma and tumor necrosis factor-alpha synergize to induce intestinal epithelial barrier dysfunction by up-regulating myosin light chain kinase expression". The American Journal of Pathology. 166 (2): 409–19. doi:10.1016/S0002-9440(10)62264-X. PMC 1237049. PMID 15681825.
- Russo JM, Florian P, Shen L, Graham WV, Tretiakova MS, Gitter AH, Mrsny RJ, Turner JR (April 2005). "Distinct temporal-spatial roles for rho kinase and myosin light chain kinase in epithelial purse-string wound closure". Gastroenterology. 128 (4): 987–1001. doi:10.1053/j.gastro.2005.01.004. PMC 1237051. PMID 15825080.
- Shimizu S, Yoshida T, Wakamori M, Ishii M, Okada T, Takahashi M, Seto M, Sakurada K, Kiuchi Y, Mori Y (January 2006). "Ca2+-calmodulin-dependent myosin light chain kinase is essential for activation of TRPC5 channels expressed in HEK293 cells". The Journal of Physiology. 570 (Pt 2): 219–35. doi:10.1113/jphysiol.2005.097998. PMC 1464317. PMID 16284075.
- Kim MT, Kim BJ, Lee JH, Kwon SC, Yeon DS, Yang DK, So I, Kim KW (April 2006). "Involvement of calmodulin and myosin light chain kinase in activation of mTRPC5 expressed in HEK cells". American Journal of Physiology. Cell Physiology. 290 (4): 1031–40. doi:10.1152/ajpcell.00602.2004. PMID 16306123.
- Connell LE, Helfman DM (June 2006). "Myosin light chain kinase plays a role in the regulation of epithelial cell survival" (PDF). Journal of Cell Science. 119 (Pt 11): 2269–81. doi:10.1242/jcs.02926. PMID 16723733. S2CID 19038438.
- Seguchi O, Takashima S, Yamazaki S, Asakura M, Asano Y, Shintani Y, Wakeno M, Minamino T, Kondo H, Furukawa H, Nakamaru K, Naito A, Takahashi T, Ohtsuka T, Kawakami K, Isomura T, Kitamura S, Tomoike H, Mochizuki N, Kitakaze M (October 2007). "A cardiac myosin light chain kinase regulates sarcomere assembly in the vertebrate heart". The Journal of Clinical Investigation. 117 (10): 2812–24. doi:10.1172/JCI30804. PMC 1978424. PMID 17885681.
- Hong F, Haldeman BD, Jackson D, Carter M, Baker JE, Cremo CR (15 June 2011). "Biochemistry of Smooth Muscle Myosin Light Chain Kinase". Archives of Biochemistry and Biophysics. 510 (2): 135–146. doi:10.1016/j.abb.2011.04.018. ISSN 0003-9861. PMC 3382066. PMID 21565153.
External links
- MYLK+protein,+human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Myosin-Light-Chain+Kinase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.